5 + 90
6 * 171
189 / 36.6
92^3
(12 + 9)^4 / 1000Introduction to R
Objectives
- Familiarize yourself with RStudio
- Learn how to work with the console
- Create and execute a script
- Create basic R objects, including vectors and data frames
Exercise Format
These exercises are in the format of a self-paced tutorial containing short explanations of key concepts, examples, and exercises for you to follow. The course uses a “learning by doing” approach, and while this first session will start with a lot of time exploring the RStudio interface, future sessions will focus heavily on having you write your own code.
Instructions for exercises will be given in the following formats:
This is a general action block. You will typically see it at the beginning of a session with instructions about the setup for that lesson.
Example: Open a blank new script and name it my_first_script.R.
This is a code block, it indicates a coding exercise where you will actually write your own code.
Example: Create an object called region that contains the value "Mandoul".
This is an observation block, it will have instructions about something that you are expected to look at or investigate.
Example: Inspect the RStudio interface.
As you move through these exercises, you may run into some errors, which occur when R is unable to complete a command. This can happen for many reasons: maybe you misspelled the name of an object, asked R to look for a file that doesn’t exist, or provided the wrong type of data to a function. Whenever an error occurs, R will stop any ongoing calculation and give you a message explaining what went wrong. Having errors is completely normal and happens to all programmers, novice and expert. Much like a natural language, R is something you will get better at the more you practice and work through your mistakes.
RStudio and R
R is a functional programming language that can be used to clean and manipulate data, run analyses (especially statistical ones), visualize results, and much more.
RStudio is a piece of software that provides a user-friendly interface for R (also called an IDE, for Integrated Development Environment). While using a graphical interface isn’t required, it is strongly recommended for beginners.
Getting Started with RStudio
Let’s get started!
Open RStudio using the start menu or desktop shortcut; if RStudio is already open, please close it and open it again.
You should see an interface that looks something like this:
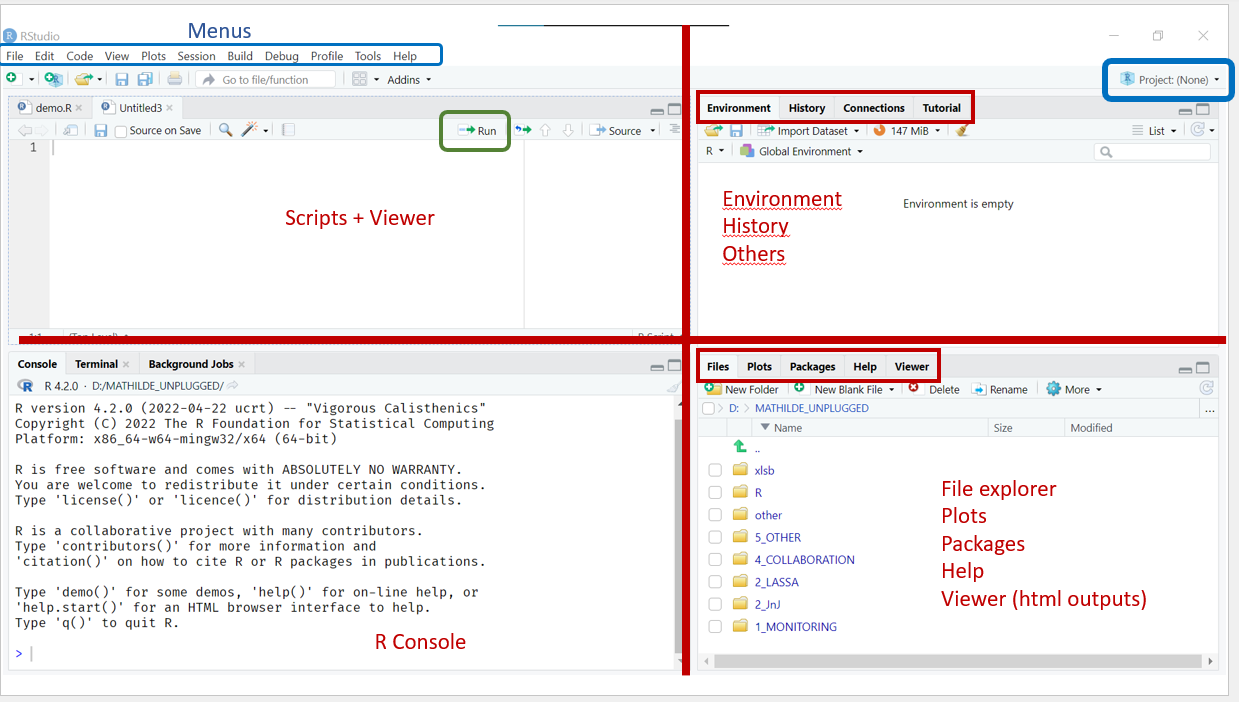
Inspect the RStudio interface.
You will have either three or four panels, including:
Upper Right Corner
To the upper right there will be a panel with several tabs. Many of these are beyond the scope of this course, but we will use the following two tabs during the course:
Environment. A list of the objects saved by the user in the current session. Because you’ve just started a new session, your environment should be empty.
History. A record of all the commands you have executed during the current session.
You can think of an R session like you would think of starting up a computer. Whenever a session starts, everything is blank and ready for computation in the same way that there aren’t any programs open when you first turn on your computer. In general, we encourage you to stop and start your R sessions regularly, you may just find that turning it off an on again will fix some of your bugs.
Bottom Right Corner
To the bottom right there will be another multi tab panel, including:
- Files. A file explorer for the working directory, which is the folder location where R is currently working.
- Plots. A location where RStudio will display static visualizations; this tab should be empty for the moment.
- Packages. A list of all the R packages installed on your computer. Packages are collections of functions that help extend the functionality of R, and we will discuss them in greater detail in the next lesson.
- Help. A place to read help pages and documentation for functions and packages.
- Viewer. A location where RStudio will display html outputs such as tables, interactive widgets, or even full on dashboards.
Left Side
- To the left (or bottom left if you have four panels) you should see the console, where R itself is run.
- To the top left (if you have four panels) will be any open scripts.
In the next two sections, let’s talk about the console and scripts in more detail.
The Console
The console is where R itself is run.
Whenever you open a new session, R will start by printing a bit of information about your set up, such as your R version number. Below this there should be a line containing the > symbol and a blinking cursor. To run a command in R, you simply need to type it in after this > and press Enter. R will then process your code and print the result (if there is one). A new > line will then appear ready for the next command.
If the last line shown in the console starts with a + instead of a > that means the console is not ready for a new command either because it is still processing a previous one or because it received a bit of incomplete code. If at any point you would like to cancel an ongoing or incomplete command, press Esc.
Run the following commands in the console one line at a time and observe the output.
Now, run the following command. Note that the final ) is missing, making the command incomplete. What happens when you do this?
3 / (2 + 97You may have noticed in the above examples that our code includes a lot of spaces between characters. This is not by accident. It is considered best practice to include spaces around most operators, such as +, -, *, /, <, >, =, and <-. Not only do these spaces make your code easier for other people to read and understand, in some (rare) cases they may even be necessary to avoid errors. That said, do be aware that there are a small number of operators that should not be surrounded by spaces, such as ^, . and :.
1+29+4.8/3*3 # BAD
1 + 29 + 4.8 / 3 * 3 # GOOD
1 ^ 2 # BAD
1^2 # GOODWe can also run functions in the console. We will discuss functions in more depth later in this lesson, but meanwhile know that the idea of functions in R is very similar to the one in Excel, where you no doubt are familiar with functions such as SUM or MEAN.
Run the following commands in the console (one line at a time).
# Find the minimum value
min(5, 10)
min(1, 8, 56, 0.3)
# Find the maximum value
max(568, 258, 314)Scripts
Scripts are text files that contain a series of commands for a particular programming language. The extension of the file indicates which language the commands were written in, and we will be using .R. Scripts allow us to create code that can be reused, shared, and even automated.
Writing Your First Script
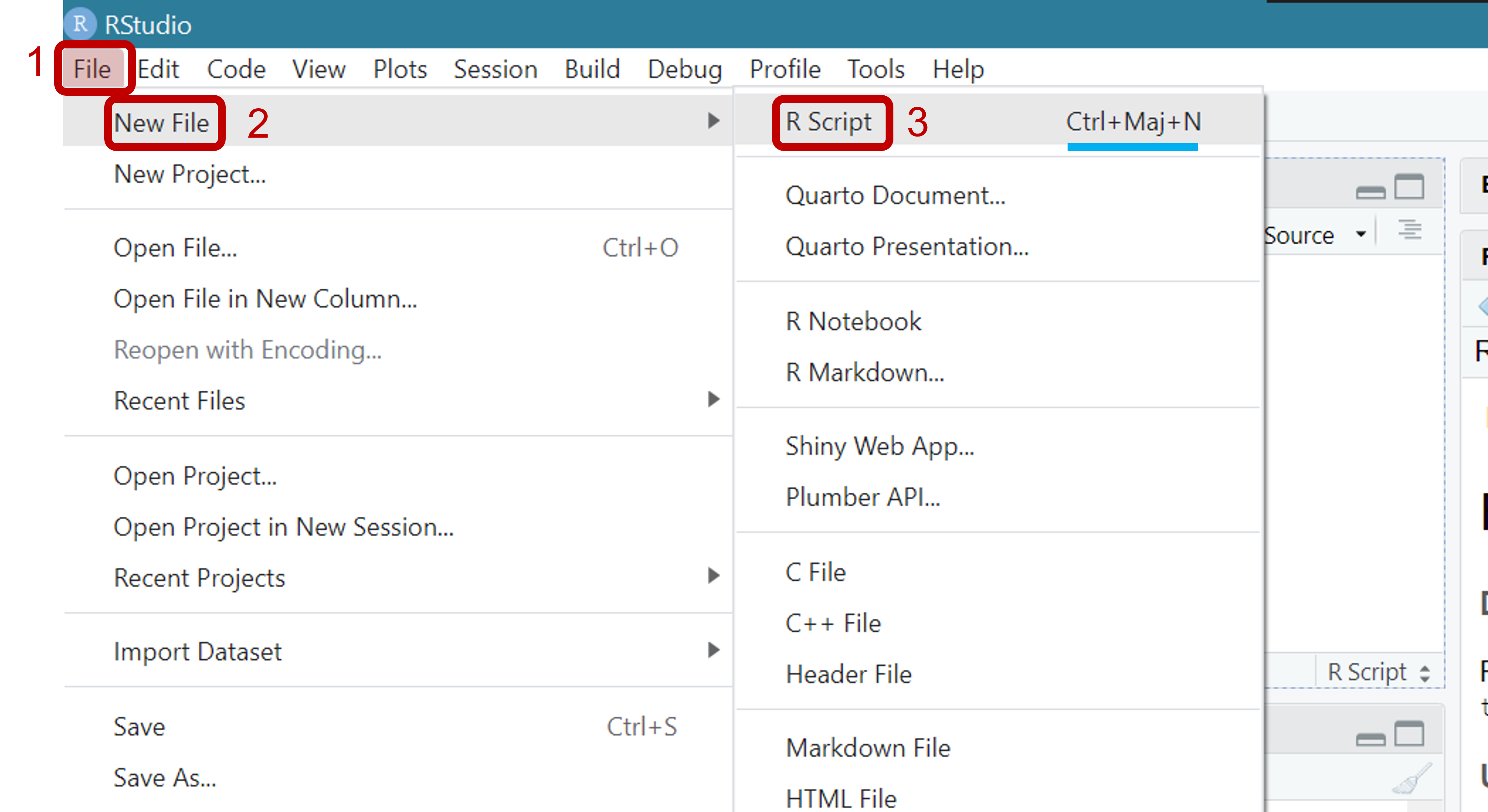
To create a new script, follow the menu File > New File > R Script. Alternatively, you can click on the small green + sign just below the File menu or use the keyboard shortcut CTRL+SHIFT+N. This new and unsaved script will appear as a blank document in the top left panel.
To save your script, either use the menu File > Save As or the keyboard shortcut CTRL+S.
Create and save a new script called discovery.R. Don’t forget to include the .R extension. For now, you can save it on your Desktop or any convenient location, but we will talk more about organizing your scripts in the next session.
Executing Code from a Script
To run code from a script simply place your cursor on the line you wish to run (or select multiple lines) and do one of the following:
- Click the
Runicon at the top right of the script panel - Use the shortcut
CTRL+Enter(cursor will move to the next line afterwards) - Use the shortcut
ALT+Enter(cursor will stay on the current line afterwards)
Copy the code you ran in the previous exercises into your script and run it using each of the above methods.
From now on, you will write your code in your script and execute it from there, unless told otherwise in the instructions.
Comments
In R, any text prefaced by a # (until the end of a line) is called a comment. R does not consider comments to be code and will ignore them whenever you run your scripts. This makes comments an excellent way to document your code.
# This is a comment
2 + 3 # This is also a commentIt is helpful to future you and others to start your scripts with a few commented lines providing some information about the file.
#### IMPORT & PREPARE DATA ####
# Author : Mathilde Mousset
# Creation Date : 23/11/2024
# Last Update : 30/11/2024
# Description : Import and clean measles surveillance data from MoissalaAdd some comments to the top of your script describing it.
Comments are also a handy way to split longer scripts into thematic sections, such as “Data Importation”, “Analysis”, “Visualization”, etc. For example:
# NAME OF SECTION 1 -----------------------------------------------
# NAME OF SECTION 2 ----------------------------------------------- Use comments to create sections in your script that correspond to the main sections in this tutorial.
Finally, comments allow us write helpful notes for our colleagues (and our future selves) that can help them understand the code and why we wrote it the way we did. The general guidance is to focus on comments that explain the “why” rather than the “what”. This is because the “what” of well written code should be relatively self explanatory.
This comment, for example, is completely superfluous:
1 + 3 # Code to add one to threeBy comparison, here are a few use cases that would warrant comments:
- You define a constant, say a seroprevalence threshold value. You may want to add a comment providing the reference for where the value comes from.
- Your code contains a value or file name that needs to be updated every week. You should indicate this with a comment to ensure that anyone else using the code is aware.
- You use a rare command or package that your colleague may not know or may find counter intuitive. You can use a comment to explain the rational behind that decision.
That being said, you are learning, and the scripts you are writing during this course are your notes, so feel free to us as many comments (of the “what” and “why” sort) as you need. You will naturally write less comments in the future, when some of the things that seem alien now become natural.
You can comment a selected line with the shortcut CTRL+SHIFT+C.
You can add a first level section with CTRL+SHIFT+R.
Add some comments to describe the code that you’ve written thus far in your script.
Data Types
R has several different data types. The ones we will see most often in this course include:
- numeric
- string (text)
- boolean (TRUE / FALSE)
- date
- factor
Numerics
The numeric type includes both integers and doubles (numbers that include a decimal) and can be created by simply writing the “naked” value into your script or console.
Strings
Strings are the R version of text and can be created by surrounding text with single or double quotation marks, for example "district" or 'cases' (double quotations are typically considered best practice).
Compare the output in the console for the following commands:
28 # numeric
"28" # text
28 + "28" # produces an errorThe last command above will give an error because we cannot perform arithmetic operations that combine text and numbers.
R is case sensitive, meaning that the string "ABC" is not the same as "abc".
If you would like to create a string that contains a quotation mark the best practice is to escape the character by putting a \ in front of it, ie: "She said \"Hello\" then left" or 'it\'s a beautiful day'. Equivalently, if you used a double quotation to create the string you can use single quotes inside of it freely (ie: "it's a beautiful day") and vice versa (i.e.: 'She said "Hello" then left').
Boolean (Logical)
The boolean (or “logical”) type stores true/false values and is created by writing either TRUE or FALSE without quotation marks.
Internally, R thinks of TRUE and FALSE as being a special version of 1 and 0 respectively, and boolean values can be easily translated to these numeric equivalents for arithmetic operations.
You may find people using T or F but this is discouraged as T and F can also be used as object or variable names. TRUE and FALSE, however, are protected in R, meaning they cannot be reassigned to another value.
Determining the Type of an Object
There are several functions to determine the type of an object (often called the class of the object in R).
Type the following commands in your script and run them:
# Get the Type of an Object
class(28)
class("Mandoul")
# Test the Type of an Object
is.numeric(28)
is.numeric("Mandoul")
is.character("Mandoul")
is.numeric(TRUE)
is.character(TRUE)
is.logical(FALSE)Creating an Object
In R, pretty much everything is an object, including functions, scalar values, and other more complex data structures. Before introducing these structures, let’s take an important detour to discuss how objects are saved into your environment.
Often, we will want to reuse the same values or data throughout a script and it is therefore very useful to store them as objects in our environment. To do this we use the assignment operator, <-.
Look at the environment panel on the top right, verifying that it is empty, then type the following command in your script and run it to save a variable called cases into your environment.
cases <- 28Look at the environment again. Is it still empty?
If you’d like to access the value of your new object, cases, you simply need to execute its name.
cases[1] 28The reason we need to wrap strings in quotation marks is actually to allow R to differentiate between strings ("cases" and object names cases).
Once created, objects can be used in other commands:
cases + 5[1] 33From your script, create an object called region that contains the value "Mandoul". Do you see it in your environment?
Don’t forget that we should always surround <- with spaces to improve readability and avoid errors.
x<-3 # BAD
x <- 3 # GOODUpdating an Object
We often want to update the value stored in an object. To do this, we simply assign a new value with the same syntax we used to create it in the first place:
cases <- 32Update the value of region to "Moyen Chari".
Object Names
When naming your objects, there are a few (relatively) hard rules:
- Don’t start with a number
- Don’t use spaces (use a
_instead) - Don’t use protected values (like
TRUEandFALSE) or function names (likemean) - Don’t use capital letters
Beyond these hard rules, there are also more subjective best practices and personal styles. In general aim for names that are short and descriptive:
a <- 19 # BAD (not informative)
age_du_patient_a_l_admission <- 19 # BAD (too long)
age <- 19 # GOODGiving your objects clear and informative names helps to make your code readable, making it easy for others to understand without the need for checking the data dictionary every two seconds.
Data Structures
Up until now we have looked only at simple objects that store single values, let’s now turn our focus to more complex structures that can store entire datasets.
Vectors
We can collect multiple values (such as numerics or strings) into a single object, called a vector.
Technically, there are several types of vectors, for example:
-
Simple vectors (or atomic vectors) can only contain one type of values. For example, a numeric vector
2, 4, 6or a string vector"Mandoul", "Moyen Chari". - Recursive vectors (usually called lists) are far more complex and can contain multiple dimensions and types of data. We will not learn about them in this lesson.
This course will not go into detail on the more abstract concepts behind these structures and instead focus only on those you will encounter most often in your daily work.
Simple Vectors
Simple vectors can contain one or more values of a single data type, they thus have two key properties: length and type. For the purpose of this class, we will use the terms “simple vector” and “vector” interchangeably (as is typical in the R community).
You’ve technically already created your first simple vector when you built cases and region. These were simply vectors with a length of one. To create a vector with more than one value, we will use the function c() (mnemonic):
cases <- c(2, 5, 8, 0, 4)Update cases with the above values and update region to create a string vector containing the values: Mandoul, Moyen-Chari, Logone Oriental, Tibesti, and Logone Occidental.
We can now use functions on the objects we have created:
mean(cases) # calculate the mean value of the cases vector[1] 3.8toupper(region) # convert all the values in region to upper case[1] "MANDOUL" "MOYEN-CHARI" "LOGONE ORIENTAL"
[4] "TIBESTI" "LOGONE OCCIDENTAL"Let’s use some functions! Try to write code that does the following:
- Calculate the sum of
casesusing the functionsum() - Convert the text in
regionto lowercase using the functiontolower()
Accessing the Values of a Vector
It is possible to access the value of a vector using square brackets containing the index (position) of the desired value, ie: [3] or [189].
cases[2] # 2nd value of cases[1] 5cases[10] # 10th value of cases[1] NAOoops it does not exist! We will come back to what this NA means in the Missing Values section.
We can also access a range of values, just as we might do in Excel. To create a range we use the : operator to separate the desired minimum and maximum index:
cases[2:4] # 2nd to 4th values of cases[1] 5 8 0Get the 3rd value of region.
Write code to access the values “Mandoul” and “Moyen-Chari” in the vector region.
Data frames
Data frames are tabular structures / 2D tables with rows and columns. It is very similar to a “table” structure in Excel. As epidemiologists, this type of data structure is perhaps the most useful and you will likely use them on a daily basis, to store linelist data for example.
Creating a data frame
We can create a data frame using the function data.frame():
data.frame(col1 = c(1, 4, 2, 9),
col2 = c("a bit of text", "some more text", "hello", "epidemiologists!")) col1 col2
1 1 a bit of text
2 4 some more text
3 2 hello
4 9 epidemiologists!See how col1 was created from a numeric vector, and col2 from a vector of strings. Here we chose the names of the columns (col1 and col2), which is the normal way, but you can run the code without to see how R handles names by default.
In your script, create a data frame called data_cases that contains cases in one column and region in the other.
Exploring a data frame
data_cases should now appear in your environment. You can click on the blue circle with a white triangle in to see some additional information, or click on its name to open the object in the same pane as the scripts to view it.
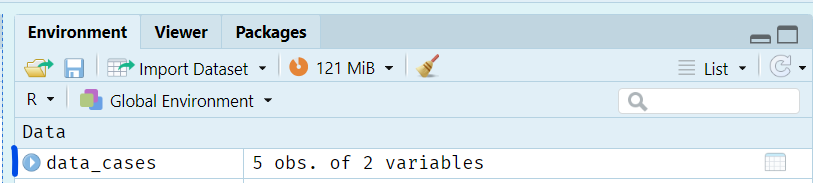
There are several handy functions we can use to explore a data frame:
Run the following commands and try to determine what type of information they are returning.
str(data_cases) # STRucture of the object
dim(data_cases) # DIMension of the object
nrow(data_cases) # Number of ROWs
ncol(data_cases) # Number of COLumns
names(data_cases) # column NAMESLet’s practice a bit more! R comes with several built in data sets that can be accessed directly, including one called iris. It is convenient today as we have not learned to import data in R yet (don’t worry, we will work on linelist data from the second session then onwards).
We can see the first few lines of this data frame using the function head():
head(iris) Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3.0 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5.0 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosaHow many rows and columns are there in iris? What are the names of its columns?
Accessing Data in a data frame
In R, there are several methods for accessing the rows and/or columns of a data frame. In this introductory session we will focus on the [ ] syntax.
We use square brackets to access single values or ranges within our data frame. To do this we must give R both a row number (or range of rows) and column number/name (or range of columns), using the syntax [row, column].
data_cases[1, 2] # the value of row one, column 2[1] "Mandoul"data_cases[1, "region"] # first value in the region column[1] "Mandoul"If we want to access all of the rows (or columns) we can simple leave a space in the place of the number/name:
data_cases[1, ] # values of all columns in row one cases region
1 2 Mandouldata_cases[2:4, ] # values of all columns for rows 2 through 4 cases region
2 5 Sud Kivu
3 8 Kasai oriental
4 0 Kasaidata_cases[ , "region"] # values of all rows for the region column[1] "Mandoul" "Sud Kivu" "Kasai oriental" "Kasai"
[5] "Haut Katanga" We can even select multiple non-consecutive indices by using a numeric vector:
data_cases[c(1, 3), ] # lines 1 and 3 (all columns) cases region
1 2 Mandoul
3 8 Kasai orientalDo be careful, as the type of output returned when extracting data from a data frame can sometimes depend on the style of indexing used:
str(data_cases[1 , ]) # returns a data frame'data.frame': 1 obs. of 2 variables:
$ cases : num 2
$ region: chr "Mandoul"str(data_cases[ , 1]) # returns a simple vector num [1:5] 2 5 8 0 4Another syntaxt to extract the various columns of a data frame:
data_cases[2] # returns the second column (as a data frame) region
1 Mandoul
2 Sud Kivu
3 Kasai oriental
4 Kasai
5 Haut Katangadata_cases["region"] # returns the region column (as a data frame) region
1 Mandoul
2 Sud Kivu
3 Kasai oriental
4 Kasai
5 Haut KatangaNotice that these commands returned single-column data frames.
Write some code to:
- extract the third value in the
regioncolumn of your data frame
- extract the second and third values of the
casescolumn
- calculate the sum of the
casescolumn of your data frame
Missing Values
As epidemiologists, we work with missing data all the time. In R, missing values are coded using a special value: NA (meaning Not Available). NA is somewhat unique in R as it doesn’t per se have a fixed type, rather, it will take on the type of the values around it. For example, an NA in a numeric column will then take on the numeric type. We will discuss the idea of missing data in more depth in later sessions of the course.
Functions
Functions are objects that contain commands (instead of values) that are run whenever the function is called. You are without doubt familiar with functions in Excel such as SUM or MEAN and the idea of functions in R is exactly the same.
Most functions require some sort of input, such as a dataset or parameter. These inputs are called arguments and are normally named. For example, when we ran sum(cases), we provided the vector cases as the first (and only) argument to the function sum().
Often, a function will have a combination of both required and optional arguments. The first argument of a function is almost always required and is typically a dataset. As an obligatory and rather obvious argument, most people omit its name when calling a function; ie: i.e. people will write mean(cases) instead of mean(x = cases). Optional arguments on the other hand are usually added using their name, i.e.: mean(x = cases, na.rm = TRUE).
Optional arguments typically have default values and we only include them when we want to change their defaults (and thus change the default behavior of the function). For example, the na.rm argument of mean() determines whether R will ignore missing values when calculating a mean. The default state of the na.rm argument is FALSE, so by default, the mean performed on data with missing values will always return NA as the result:
mean(c(1, 3, NA))[1] NAThis is true for many arithmetic operations in R. If we want R to calculate the mean on whatever data is available (and ignore the missing values) we need to explicitly set na.rm = TRUE:
mean(c(1, 3, NA), na.rm = TRUE)[1] 2Notice that arguments are separated by commas. These commas should always be followed by a space and whenever a named argument is used the = should be surrounded by spaces:
mean(cases,na.rm=TRUE) # BAD
mean(cases, na.rm = TRUE) # GOODAs you work with increasingly complex functions, you may start to have a lot of arguments. For readability, it is typically recommended to split each argument onto its own line:
mean(cases,
na.rm = TRUE) What happens if we put the arguments in the wrong order? If you provided the name of the arguments in you command, the function will still work exactly as expected. That being said, doing this would make your code harder to read and we encourage you to stick with a standard order of putting obligatory arguments like data first.
# technically functional but hard to read:
mean(na.rm = TRUE,
x = cases)
# better:
mean(cases,
na.rm = TRUE)Of course, if you mess up the ordering of arguments and didn’t include their names your code will not work as expected, or even throw an error:
mean(TRUE, cases) # not what you expectDone!
That’s all for this session, congratulations on taking your first steps with R and RStudio!